Data Acquisition Framework Using Custom Python Wrapper For Concurrent BCP Utility Execution
Although building a data acquisition framework for a data warehouse isn’t nearly or as interesting as doing analytics or data mining on the already well-structured data, it’s an essential part of any data warehousing project. I have already written extensively on how one could build a robust data acquisition framework in THIS series of blog posts, however, given the rapidly growing ecosystem of data sources and data stores, more and more projects tend to gravitate to the more application-agnostic storage formats e.g. text files or column-oriented files e.g. parquet. As such, direct data store connection may not be a possibility or the most optimal way of exporting data, making flat file export/import option a good alternative.
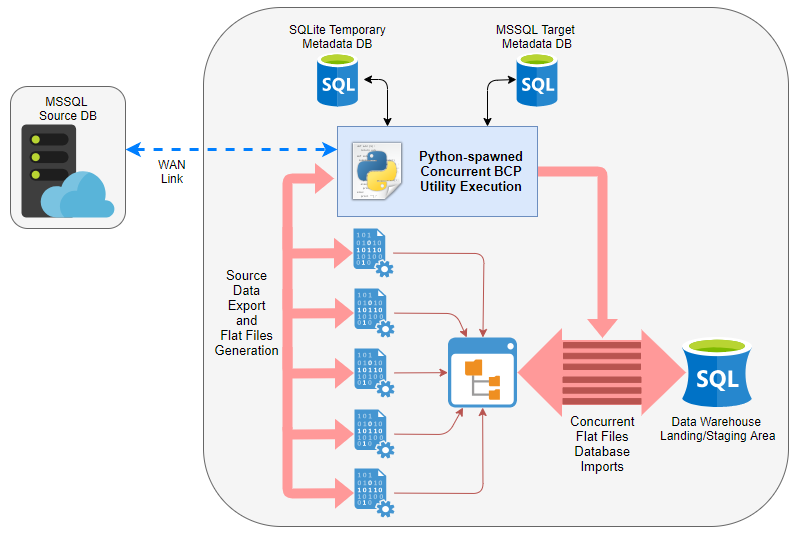
Microsoft offers a few different tools to facilitate flat files imports and exports, the most commonly used being bcp, BULK INSERT and their own ETL tool, capable of connecting to and reading flat files – SSIS. In this demo, I will analyse the difference in performance for data export and import between using pure T-SQL (executed as a series of concurrently running SQL Agent jobs) and parallelised bcp operations (using custom Python wrapper). Below image depicts high-level architecture of the solution with a short video clip demonstrating the actual code execution on a sample dataset at the end of this post. Let’s compare the pair and see if any of them is better at moving data across the wire.
Database and Schema Setup
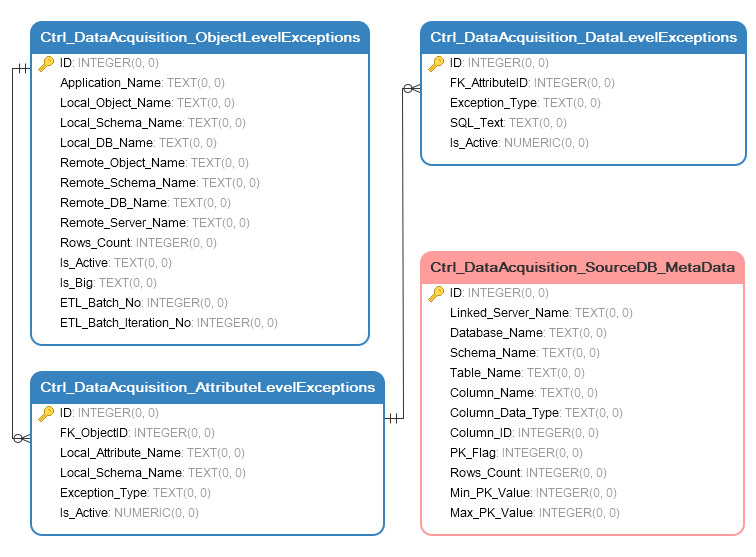
Most of data warehouses come with some form of metadata layer so for this exercise I have ‘scrapped’ the source metadata e.g. attributes such as object names, data types, row counts etc. into a Sqlite local database. I also copied over partial target database metadata (with a bit more structure imposed) from one of the systems I’m currently working with. This resulted in creating the following schema of tables containing key information on source and target objects, their attributes, their state etc.
Target metadata is contained to three tables which allow for a three-layer, hierarchical approach to data acquisition:
- Ctrl_DataAcquisition_ObjectLevelExceptions table stores details of database objects with some other peripheral information. The grain of this table is set to the object (table) level which then allows for an easy object tagging and acquisitions inclusion/exclusion.
- Ctrl_DataAcquisition_AttributeLevelExceptions table stores attributes level data. It references ‘objects’ table and determines which fields out of the referenced object are to be included/excluded.
- Ctrl_DataAcquisition_DataLevelExceptions table stores any statements which need to be executed in order to remove/retain specific values.
As a result, thanks to this setup, we now have the option to dictate which objects are to be acquired, which fields from those objects are to be retained/omitted and finally which values out of these attributes are to be persisted/hashed/obfuscated/removed etc. Source-level metadata (in this scenario) was diluted to a single table containing some of the key attributes and data to fulfil the requirements for this post.
Creating the metadata repository, depending on the system’s complexity and level of detail required, can be quite challenging in itself so I will leave out the particulars out of this post, however, it’s important to note that this mini-framework would not be possible without storing and comparing both systems’ metadata i.e. source database and target database.
This post also scratches the surface of actual metadata layer implementation details but suffice to say that any well designed and architected framework should lean heavily on metadata. For this post, mainly to preserve the brevity and the focus on the problem analysed, I have obfuscated the details of target metadata creation and diluted source metadata ‘scrapping’ to bare minimum.
The key information sourced, aside from which tables and columns to handle, is essential to providing a more flexible approach to data extraction for the following reasons:
- Primary key flag – identifies a specific attribute as either a primary key or not a primary key in order to split a large table into multiple ‘chunks’. As most primary keys are of data type INTEGER (or its derivative e.g. SMALLINT, TINYINT, BIGINT) splitting larger tables into smaller streams of data and files allows for better resources utilisation.
- Column data type – useful in checking whether arithmetic can be performed on the given table’s primary key i.e. if it’s not an INTEGER (or similar) than subdividing into smaller partitions will not be an option.
- Rows count – this data is required I order to partition larger tables into smaller shards based on the total record count.
- Minimum record count – this value is essential for partitioning larger tables which do not have their seeding values starting from one.
- Maximum record count – same as above, this piece of data is required to ascertain the ceiling value for further data ‘chunking’.
The following Python script was used to create and populate MetadataDB.db database.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 | import configparserimport sqlite3import osimport sysimport pyodbcconfig = configparser.ConfigParser()config.read("params.cfg")dbsqlite_location = config.get("Sqlite", os.path.normpath("dbsqlite_location"))dbsqlite_fileName = config.get("Sqlite", "dbsqlite_fileName")dbsqlite_sql = config.get("Sqlite", "dbsqlite_sql")dbsqlite_import_tables = config.get("Tables_To_Import", "local_mssql_meta_tables")dbsqlite_import_tables = dbsqlite_import_tables.split(",")dbsqlite_excluded_attribs = config.get("Tables_To_Import", "local_mssql_meta_attributes_to_exclude")dbsqlite_excluded_attribs = dbsqlite_excluded_attribs.split(",")dbsqlite_import_tables_remote = config.get("Tables_To_Import", "remote_meta_tables")def get_sql(dbsqlite_location, dbsqlite_fileName, dbsqlite_sql): """ Source operation types from the SQL file - denoted by the use of four dash characteres and store them in a dictionary (referenced later) """ if os.path.isfile(dbsqlite_fileName): os.remove(dbsqlite_fileName) operations = [] commands = [] with open(dbsqlite_sql, "r") as f: for i in f: if i.startswith("----"): i = i.replace("----", "") operations.append(i.rstrip("\n")) tempCommands = [] with open(dbsqlite_sql, "r") as f: for i in f: tempCommands.append(i) l = [i for i, s in enumerate(tempCommands) if "----" in s] l.append((len(tempCommands))) for first, second in zip(l, l[1:]): commands.append("".join(tempCommands[first:second])) sql = dict(zip(operations, commands)) return sqldef build_db(dbsqlite_location, dbsqlite_fileName, sql): """ Build the database """ print("Building TableauEX.db database schema... ") with sqlite3.connect( os.path.join(dbsqlite_location, dbsqlite_fileName) ) as sqlLiteConn: sqlCommands = sql.get("SQL 1: Build_Meta_DB").split(";") for c in sqlCommands: try: sqlLiteConn.execute(c) except Exception as e: print(e) sqlLiteConn.rollback() sys.exit(1) else: sqlLiteConn.commit()def populate_db_from_mssql_meta( dbsqlite_import_tables, dbsqlite_excluded_attribs, sql, **options): """ Source local instance MSSQL metadata and populate Sqlite tables with the data. """ mssql_args = { "Driver": options.get("Driver", "ODBC Driver 13 for SQL Server"), "Server": options.get("Server", "ServerName\\InstanceName"), "Database": options.get("Database", "Metadata_DB_Name"), "Trusted_Connection": options.get("Trusted_Connection", "yes"), } cnxn = pyodbc.connect(**mssql_args) if cnxn: with sqlite3.connect( os.path.join(dbsqlite_location, dbsqlite_fileName) ) as sqlLiteConn: mssql_cursor = cnxn.cursor() for tbl in dbsqlite_import_tables: try: tbl_cols = {} mssql_cursor.execute( """SELECT c.ORDINAL_POSITION, c.COLUMN_NAME FROM Metadata_DB_Name.INFORMATION_SCHEMA.COLUMNS c WHERE c.TABLE_SCHEMA = 'dbo' AND c.TABLE_NAME = '{}' AND c.COLUMN_NAME NOT IN ({}) ORDER BY 1 ASC;""".format( tbl, ", ".join( map(lambda x: "'" + x + "'", dbsqlite_excluded_attribs) ), ) ) rows = mssql_cursor.fetchall() for row in rows: tbl_cols.update({row[0]: row[1]}) sortd = [tbl_cols[key] for key in sorted(tbl_cols.keys())] cols = ",".join(sortd) mssql_cursor.execute("SELECT {} FROM {}".format(cols, tbl)) rows = mssql_cursor.fetchall() num_columns = max(len(rows[0]) for t in rows) mssql = "INSERT INTO {} ({}) VALUES({})".format( tbl, cols, ",".join("?" * num_columns) ) sqlLiteConn.executemany(mssql, rows) except Exception as e: print(e) sys.exit(1) else: sqlLiteConn.commit()def populate_db_from_meta(dbsqlite_import_tables_remote, sql, **options): """ Source remote MSSQL server metadata and populate nominated Sqlite table with the data. """ mssql_args = { "Driver": options.get("Driver", "ODBC Driver 13 for SQL Server"), "Server": options.get("Server", "ServerName\\InstanceName"), "Database": options.get("Database", "Metadata_DB_Name"), "Trusted_Connection": options.get("Trusted_Connection", "yes"), } with sqlite3.connect( os.path.join(dbsqlite_location, dbsqlite_fileName) ) as sqlLiteConn: print("Fetching external SQL Server metadata... ") cnxn = pyodbc.connect(**mssql_args) if cnxn: mssql_cursor = cnxn.cursor() sqlCommands = sql.get("SQL 2: Scrape_Source_DH2_Meta") try: mssql_cursor.execute(sqlCommands) rows = mssql_cursor.fetchall() num_columns = max(len(rows[0]) for t in rows) mssql = "INSERT INTO {} (linked_server_name, \ database_name, \ schema_name, \ table_name, \ column_name, \ column_data_type, \ column_id, \ pk_flag, \ rows_count, \ min_pk_value, \ max_pk_value) \ VALUES({})".format( dbsqlite_import_tables_remote, ",".join("?" * num_columns) ) sqlLiteConn.executemany(mssql, rows) except Exception as e: sys.exit(1) else: sqlLiteConn.commit()def main(): sql = get_sql(dbsqlite_location, dbsqlite_fileName, dbsqlite_sql) build_db(dbsqlite_location, dbsqlite_fileName, sql) populate_db_from_mssql_meta(dbsqlite_import_tables, dbsqlite_excluded_attribs, sql) populate_db_from_meta(dbsqlite_import_tables_remote, sql)if __name__ == '__main__': main() |
At runtime, this script also calls a SQL query which copies data from an already existing metadata database (on the target server) as well as queries source database for all of its necessary metadata. The full SQL file as well as all other scripts used in this post can be downloaded from my OneDriver folder HERE. When populated with the source and target data, a SQL query is issued as part of the subsequent script execution which joins source and target metadata and ‘flattens’ the output, comma-delimiting attribute values (most column names blurred-out) into an array used to specify individual columns names as per the screenshot below (click on image to enlarge).
Python Wrapper For Concurrent BCP Utility Execution
Now that we have our source and target metadata defined we can start testing transfer times. In order to take advantage of all resources available, multiple bcp processes can be spawned using Python’s multiprocessing library for both: extracting data out of the source environment as well as importing it into target database. The following slapdash Python script, comprised mainly of the large SQL statement which pulls source and target metadata together, can be used to invoke concurrent bcp sessions and further subdivide larger tables into smaller batches which are streamed independently of each other.
The script, along with the database structure, can be further customised to include features such as allowing incremental, delta-only data transfer and error logging via either Python-specific functionality or bcp utility ‘-e err_file’ switch usage with just a few lines of code.
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 138 139 140 141 142 143 144 145 146 147 148 149 150 151 152 153 154 155 156 157 158 159 160 161 162 163 164 165 166 167 168 169 170 171 172 173 174 175 176 177 178 179 180 181 182 183 184 185 186 187 188 189 190 191 192 193 194 195 196 197 198 199 200 201 202 203 204 205 206 207 208 209 210 211 212 213 214 215 216 217 218 219 220 221 222 223 224 225 226 227 228 229 230 231 232 233 234 235 236 237 238 239 240 241 242 243 244 245 246 247 248 249 250 251 252 253 254 255 256 257 258 259 260 261 262 263 264 265 266 267 268 269 270 271 272 273 274 275 276 277 278 279 280 281 282 283 284 285 286 287 288 289 290 291 292 293 294 295 296 297 298 299 300 301 302 303 304 305 306 307 308 309 310 311 312 313 314 315 316 317 318 319 320 321 322 323 324 325 326 327 328 329 330 331 332 333 334 335 336 337 338 339 340 341 342 343 344 345 346 347 348 349 350 351 352 353 354 355 356 357 358 359 360 361 362 363 364 365 366 367 368 369 370 371 372 373 374 375 376 377 378 379 380 381 382 383 384 385 386 387 388 389 390 391 392 | from os import path, systemfrom multiprocessing import Pool, cpu_countimport argparseimport timeimport pyodbcimport sqlite3import configparserimport build_dbodbcDriver = "{ODBC Driver 13 for SQL Server}"config = configparser.ConfigParser()config.read("params.cfg")dbsqlite_location = config.get("Sqlite", path.normpath("dbsqlite_location"))dbsqlite_fileName = config.get("Sqlite", "dbsqlite_fileName")dbsqlite_sql = config.get("Sqlite", "dbsqlite_sql")export_path = config.get("Files_Path", path.normpath("export_path"))odbcDriver = "{ODBC Driver 13 for SQL Server}"parser = argparse.ArgumentParser( description="SQL Server Data Loading/Extraction BCP Wrapper by bicortex.com")parser.add_argument( "-SSVR", "--source_server", help="Source Linked Server Name", required=True, type=str)parser.add_argument( "-SDB", "--source_database", help="Source Database Name", required=True, type=str)parser.add_argument( "-SSCH", "--source_schema", help="Source Schema Name", required=True, type=str)parser.add_argument( "-TSVR", "--target_server", help="Target Server Name", required=True, type=str)parser.add_argument( "-TDB", "--target_database", help="Target Database Name", required=True, type=str)parser.add_argument( "-TSCH", "--target_schema", help="Target Schema Name", required=True, type=str)args = parser.parse_args()if ( not args.source_server or not args.source_database or not args.source_schema or not args.target_server or not args.target_database or not args.target_schema): parser.print_help() exit(1)def get_db_data(linked_server_name, remote_db_name, remote_schema_name): """ Connect to Sqlite database and get all metadata required for sourcing target data and subsequent files generation """ with sqlite3.connect( path.join(dbsqlite_location, dbsqlite_fileName) ) as sqlLiteConn: sqliteCursor = sqlLiteConn.cursor() sqliteCursor.execute( """ SELECT Database_Name, Schema_Name, Table_Name, GROUP_CONCAT( Column_Name ) AS Column_Names, Is_Big, ETL_Batch_No, Rows_Count, Min_PK_Value, Max_PK_Value, GROUP_CONCAT( CASE WHEN PK_Column_Name != 'N/A' THEN PK_Column_Name END) AS PK_Column_Name FROM (SELECT b.Remote_DB_Name AS Database_Name, b.Remote_Schema_Name AS Schema_Name, a.Table_Name, a.Column_Name, b.Is_Big, b.ETL_Batch_No, b.Rows_Count, CASE WHEN a.PK_Flag = 1 THEN a.Column_Name ELSE 'N/A' END AS PK_Column_Name, a.Min_PK_Value, a.Max_PK_Value FROM Ctrl_DataAcquisition_SourceDB_MetaData a JOIN Ctrl_DataAcquisition_ObjectLevelExceptions b ON a.Table_Name = b.Remote_Object_Name AND b.Remote_Schema_Name = '{schema}' AND b.Remote_DB_Name = '{db}' AND b.Remote_Server_Name = '{lsvr}' WHERE b.Is_Active = 1 AND a.Column_Data_Type != 'timestamp' AND a.Rows_Count > 0 AND (Min_PK_Value != -1 OR Max_PK_Value != -1) AND NOT EXISTS( SELECT 1 FROM Ctrl_DataAcquisition_AttributeLevelExceptions o WHERE o.Is_Active = 1 AND o.Local_Attribute_Name = a.Column_Name AND o.FK_ObjectID = b.ID ) ORDER BY 1, 2, 3, a.Column_ID) GROUP BY Database_Name, Schema_Name, Table_Name, Is_Big, ETL_Batch_No, Rows_Count, Min_PK_Value """.format( lsvr=linked_server_name, schema=remote_schema_name, db=remote_db_name ) ) db_data = sqliteCursor.fetchall() return db_datadef split_into_ranges(start, end, parts): """ Based on input parameters, split number of records into consistent and record count-like shards of data and return to the calling function """ ranges = [] x = round((end - start) / parts) for _ in range(parts): ranges.append([start, start + x]) start = start + x + 1 if end - start <= x: remainder = end - ranges[-1][-1] ranges.append([ranges[-1][-1] + 1, ranges[-1][-1] + remainder]) break return rangesdef truncate_target_table(schema, table, **options): """ Truncate target table before data is loaded """ mssql_args = {"Driver": options.get("Driver", "ODBC Driver 13 for SQL Server"), "Server": options.get("Server", "ServerName\\InstanceName"), "Database": options.get("Database", "Staging_DB_Name"), "Trusted_Connection": options.get("Trusted_Connection", "yes"), } cnxn = pyodbc.connect(**mssql_args) if cnxn: try: mssql_cursor = cnxn.cursor() sql_truncate = "TRUNCATE TABLE {db}.{schema}.{tbl};".format( db=mssql_args.get("Database"), schema=schema, tbl=table) print("Truncating {tbl} table...".format(tbl=table)) mssql_cursor.execute(sql_truncate) cnxn.commit() sql_select = "SELECT TOP 1 1 FROM {db}.{schema}.{tbl};".format( db=mssql_args.get("Database"), schema=schema, tbl=table) mssql_cursor.execute(sql_select) rows = mssql_cursor.fetchone() if rows: raise Exception( "Issue truncating target table...Please troubleshoot!") except Exception as e: print(e) finally: cnxn.close()def export_import_data(target_server, target_database, target_schema, source_server, source_database, source_schema, table_name, columns, export_path, ): """ Export source data into a 'non-chunked' CSV file calling SQL Server bcp utility and import staged CSV file into the target SQL Server instance. This function also provides a verbose way to output runtime details of which object is being exported/imported and the time it took to process. """ full_export_path = path.join(export_path, table_name + ".csv") start = time.time() print("Exporting '{tblname}' table into {file} file...".format( tblname=table_name, file=table_name + ".csv")) bcp_export = 'bcp "SELECT * FROM OPENQUERY ({ssvr}, \'SELECT {cols} FROM {sdb}.{schema}.{tbl}\')" queryout {path} -T -S {tsvr} -q -c -t "|" -r "\\n" 1>NUL'.format( ssvr=source_server, sdb=source_database, schema=source_schema, cols=columns, tbl=table_name, path=full_export_path, tsvr=target_server, ) system(bcp_export) end = time.time() elapsed = round(end - start) if path.isfile(full_export_path): print("File '{file}' exported in {time} seconds".format( file=table_name + ".csv", time=elapsed )) start, end, elapsed = None, None, None start = time.time() print( "Importing '{file}' file into {tblname} table...".format( tblname=table_name, file=table_name + ".csv" ) ) bcp_import = 'bcp {schema}.{tbl} in {path} -S {tsvr} -d {tdb} -h "TABLOCK" -T -q -c -t "|" -r "\\n" 1>NUL'.format( schema=target_schema, tbl=table_name, path=full_export_path, tsvr=target_server, tdb=target_database, ) system(bcp_import) end = time.time() elapsed = round(end - start) print("File '{file}' imported in {time} seconds".format( file=table_name + ".csv", time=elapsed ))def export_import_chunked_data( target_server, target_database, target_schema, source_server, source_database, source_schema, table_name, columns, export_path, vals, idx, pk_column_name,): """ Export source data into a 'chunked' CSV file calling SQL Server bcp utility and import staged CSV file into the target SQL Server instance. This function also provides a verbose way to output runtime details of which object is being exported/imported and the time it took to process. """ full_export_path = path.join(export_path, table_name + str(idx) + ".csv") start = time.time() print( "Exporting '{tblname}' table into {file} file ({pk}s between {minv} and {maxv})...".format( tblname=table_name, file=table_name + str(idx) + ".csv", pk=pk_column_name, minv=str(int(vals[0])), maxv=str(int(vals[1])), ) ) bcp_export = 'bcp "SELECT * FROM OPENQUERY ({ssvr}, \'SELECT {cols} FROM {sdb}.{schema}.{tbl} WHERE {pk} BETWEEN {minv} AND {maxv}\')" queryout {path} -T -S {tsvr} -q -c -t "|" -r "\\n" 1>NUL'.format( ssvr=source_server, sdb=source_database, schema=source_schema, cols=columns, tbl=table_name, path=full_export_path, tsvr=target_server, pk=pk_column_name, minv=str(int(vals[0])), maxv=str(int(vals[1])), ) system(bcp_export) end = time.time() elapsed = round(end - start) if path.isfile(full_export_path): print("File '{file}' exported in {time} seconds".format( file=table_name + str(idx) + ".csv", time=elapsed )) start, end, elapsed = None, None, None start = time.time() print( "Importing '{file}' file into {tblname} table ({pk}s between {minv} and {maxv})...".format( tblname=table_name, file=table_name + str(idx) + ".csv", pk=pk_column_name, minv=str(int(vals[0])), maxv=str(int(vals[1])), ) ) bcp_import = 'bcp {schema}.{tbl} in {path} -S {tsvr} -d {tdb} -T -q -c -t "|" -r "\\n" 1>NUL'.format( schema=target_schema, tbl=table_name, path=full_export_path, tsvr=target_server, tdb=target_database, ) system(bcp_import) end = time.time() elapsed = round(end - start) print("File '{file}' imported in {time} seconds".format( file=table_name + str(idx) + ".csv", time=elapsed ))def main(): """ Call export/import 'chunked' and 'non-chunked' data functions and process each database object as per the metadata information in a concurrent fashion """ build_db.main() db_data = get_db_data(args.source_server, args.source_database, args.source_schema) if db_data: if path.exists(export_path): try: p = Pool(processes=cpu_count()) for row in db_data: table_name = row[2] columns = row[3] is_big = int(row[4]) etl_batch_no = int(row[5]) min_pk_value = int(row[7]) max_pk_value = int(row[8]) pk_column_name = row[9] truncate_target_table( schema=args.target_schema, table=table_name) if is_big == 1: ranges = split_into_ranges( min_pk_value, max_pk_value, etl_batch_no ) for idx, vals in enumerate(ranges): p.apply_async( export_import_chunked_data, [ args.target_server, args.target_database, args.target_schema, args.source_server, args.source_database, args.source_schema, table_name, columns, export_path, vals, idx, pk_column_name, ], ) else: p.apply_async( export_import_data, [ args.target_server, args.target_database, args.target_schema, args.source_server, args.source_database, args.source_schema, table_name, columns, export_path, ], ) p.close() p.join() except Exception as e: print(e) else: raise Exception( "Specyfied folder does not exist. Please troubleshoot!") else: raise Exception( "No data retrieved from the database...Please troubleshoot!")if __name__ == "__main__": main() |
Once kicked off, the multiprocessing Python module spins up multiple instances of bcp utility. The below footage demonstrates those running as individual processes (limited to 4 based on the number of cores allocated to the test virtual machine) on data transfer limited to 4 test objects: tables named ‘client’, ‘clientaddress’, ‘news’, ‘report’. Naturally, the more cores allocated the better the performance should be.
Testing Results
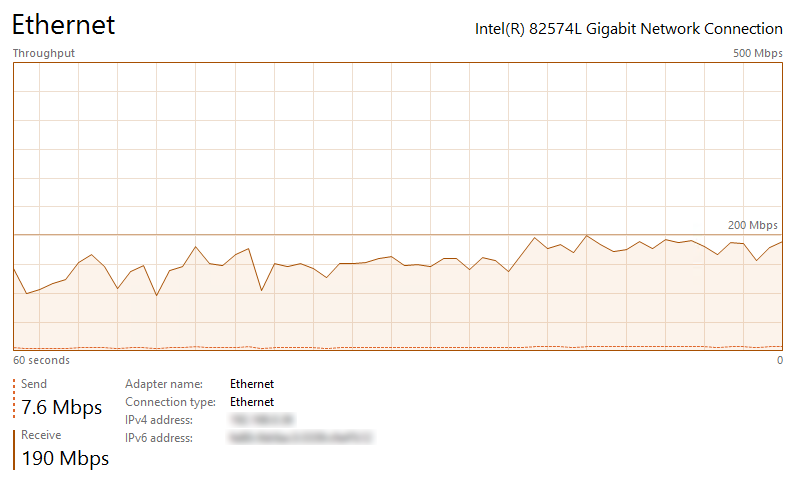
Running the script with a mixture of different tables (60 in total), across a 15Mbps link, on a 4 core machine with 64GB DDR4 RAM allocated to the instance (target environment) yielded pretty much the same transfer speeds results as using a direct RDBMS-to-RDBMS connection. The volume of data (table size – index size) equaling to around 7 GB in SQL Server (4GB as extracted CSV files) yielded a disappointing 45 minutes transfer time. The key issue with my setup specifically was the mediocre network speed, with SQL Server engine having to spend around 75 percent of this time (33 minutes) waiting for data to arrive. Upgrading network link connection allowed for much faster processing time, making this method of data acquisition and staging a very compelling option for a contingency or fail-over method of sourcing application data into landing area of a data warehouse environment. Below images show how upgrading network speed (most of times the new connection speed oscillated between 140Mbps and 160Mbps due to VPN/firewall restrictions) removed the network bottleneck and maximized hardware resources utilization across this virtual environment (vCPUs usage no longer idling, waiting for network packets).
When re-running the same acquisition across the upgraded link the processing time was reduced down to 5 minutes.
So there you go, a simple and straightforward micro-framework for concurrent bcp utility data extraction which can be used as a contingency method for data acquisitions or for testing purposes.
http://scuttle.org/bookmarks.php/pass?action=addThis entry was posted on Tuesday, October 2nd, 2018 at 8:10 am and is filed under Programming, SQL, SQL Server. You can follow any responses to this entry through the RSS 2.0 feed. You can leave a response, or trackback from your own site.




Never thought Polybase could be used in this capacity and don't think Microsoft advertised this feature (off-loading DB data as…