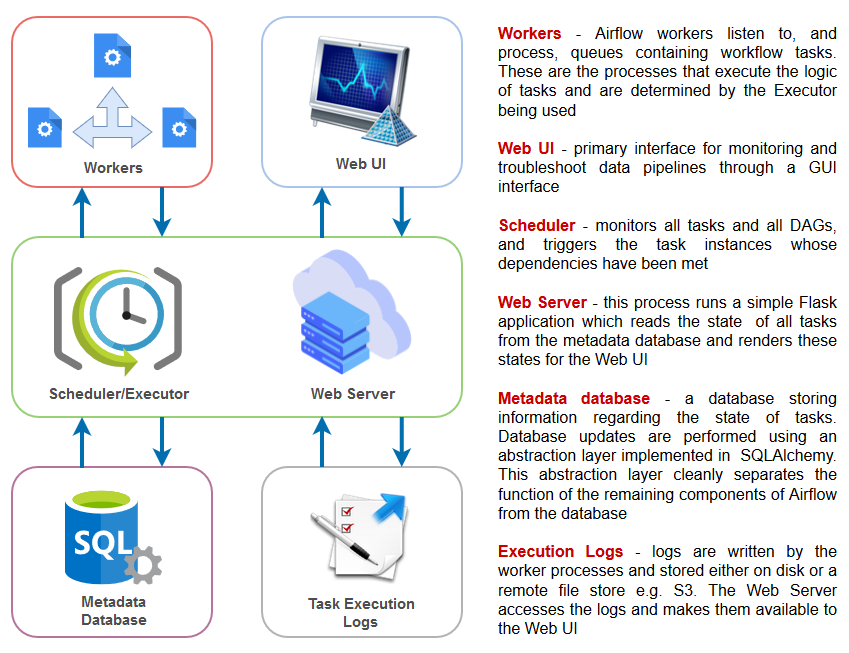
Kicking the Tires on Airflow, Apache’s workflow management platform – Architecture Overview, Installation and sample Azure Cloud Deployment Pipeline in Python (Part 2)
May 24th, 2020 / 4 Comments » / by admin
Airflow Sample Workflow Continued…
Note: Part 1 can be found HERE and all files used in this tutorial can be downloaded from HERE.
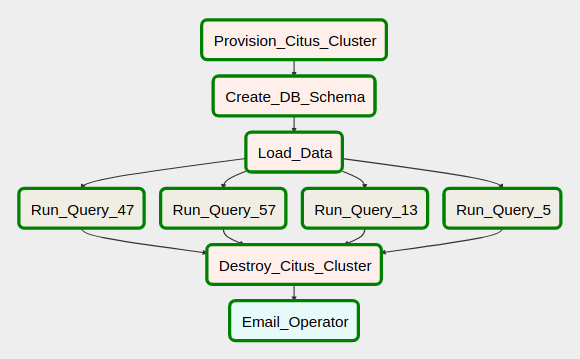
In the first part to this short series on Apache Airflow I outlined it its core architecture, installation process and created a sample example of a very rudimentary DAG (Directed Acyclic Graph). In this post I’d like to dive a bit deeper and look at a more advanced workflow which combines a few different operations and tasks. Let’s assume that we’re tasked with creating a DAG which needs to accomplish the following:
- Programmatically create Citus database instance in Azure public cloud with specific configuration parameters e.g. geographic location, storage size, number of nodes assigned, firewall rules etc. These parameters are stored across two JSON template files, known Azure Resource Manager templates
- Subsequently to creating Azure Citus database, the workflow should create a database schema using DDL SQL stored in one of the configuration files
- Next, load text files data into the newly created database schema
- Run four sample TPC-DS SQL queries in parallel (queries 5, 13, 47 and 57) and store the results into a file on the local file system
- Shut the cluster down and remove any resources created as part of this process
- Send out an e-mail notifying operator(s) of the workflow conclusion
A similar type of workload is often used to create an idempotent pipeline which stitches together a number of tasks, including cloud infrastructure provisioning, data analytics, ETL/ELT activities etc. to design an end-to-end data management solution. The required DAG, with all its dependencies and activities should like as per the image below.
Sample Airflow Azure Cloud Pipeline Architecture
In order to provision the Azure resource groups and resources, I have created two JSON template files, also known as Azure Resource Manager (ARM) templates, which contain the required resources and their properties. ARM templates allow for declarative creation and deployment of any Azure infrastructure components e.g. virtual machines, hosted databases, storage accounts etc. along their properties and names. As the libraries used in the Python version of the Azure SDK directly mirror and consume Azure service’s REST endpoints, it’s quite easy to script out a repeatable deployment process using ARM templates as described in more details further along in this post. As part of the solution configuration, I have also created two SQL files which are used to create a suite of tables on the Citus cluster and run the queries, outputting the result sets into a local directory.
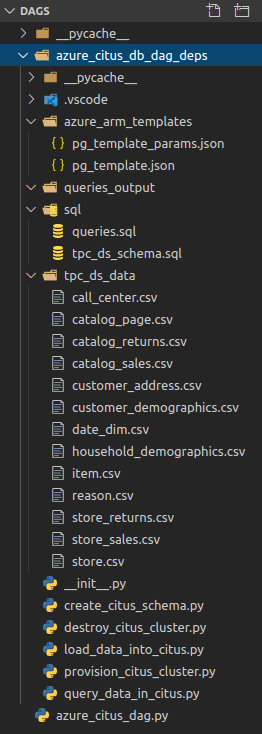 The aforementioned files used to populate the Citus cluster schema are part of the TPC-DS benchmark data (scaling factor set to 1GB) and consist of a number of flat files in a CSV format. As we’re not going to recreate the whole TPC-DS benchmark suite of tests (this post is not intended to evaluate Citus database performance), to simplify the data loading process, I will only use a few of them (just over 800MB in size), which should provide us with enough data to run a few queries once the tables have been populated. To simplify this demonstration, all Python solution files as well as ARM JSON templates, SQL files and flat files data were moved into dags directory aka {AIRFLOW_HOME}/dags folder. Under typical scenario, data would not co-exists with configuration and solution files but for this exercise, I have nominated to structure the project as per the the directory breakdown on the left. Also, in a production environment, particularly one with many dag and task files, you would want to ensure that a consistent organizational structure is imposed across DAG folder and its sub-directories. As Airflow is such a versatile tool, depending on a specific use case, you may want to separate individual projects and their related hooks, operators, scripts and other files into their own respective folders etc.
The aforementioned files used to populate the Citus cluster schema are part of the TPC-DS benchmark data (scaling factor set to 1GB) and consist of a number of flat files in a CSV format. As we’re not going to recreate the whole TPC-DS benchmark suite of tests (this post is not intended to evaluate Citus database performance), to simplify the data loading process, I will only use a few of them (just over 800MB in size), which should provide us with enough data to run a few queries once the tables have been populated. To simplify this demonstration, all Python solution files as well as ARM JSON templates, SQL files and flat files data were moved into dags directory aka {AIRFLOW_HOME}/dags folder. Under typical scenario, data would not co-exists with configuration and solution files but for this exercise, I have nominated to structure the project as per the the directory breakdown on the left. Also, in a production environment, particularly one with many dag and task files, you would want to ensure that a consistent organizational structure is imposed across DAG folder and its sub-directories. As Airflow is such a versatile tool, depending on a specific use case, you may want to separate individual projects and their related hooks, operators, scripts and other files into their own respective folders etc.
With Airflow server up and running (installation and configuration instructions in the previous post HERE) and the files required to load into our Citus database schema staged in the solution directory we can start unpacking individual code samples used in the final DAG. As mentioned at the start, Citus database cluster, just as any other Azure resource, can be spun up declaratively using ARM template(s). You can peruse the content of the templates used in this demo as well as other artifacts in my publicly accessible OneDrive folder HERE.
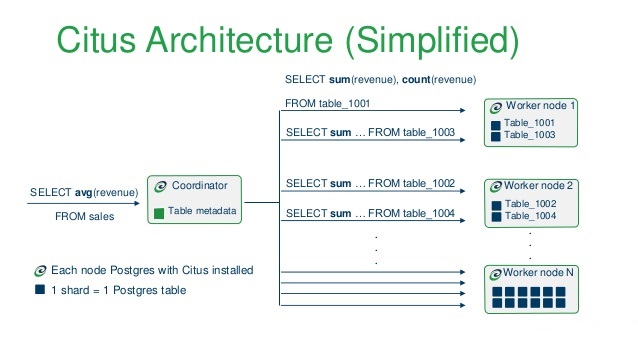
A quick word about Citus (now owned by Microsoft). Citus is an open source extension to PostgreSQL which distributes data and queries across multiple nodes. Because Citus is an extension to PostgreSQL (not a fork), it gives developers and enterprises a scale-out database while keeping the power and familiarity of a relational database. Citus horizontally scales PostgreSQL across multiple machines using sharding and replication. Its query engine parallelizes incoming SQL queries across these servers to enable super-fast responses on even very large datasets. I’m not very familiar with the service but there are many posts on the Internet describing some interesting projects Citus was used in, beating other competing technologies (TiDB, Apache Pinot, Apache Kylin, Apache Druid), partly due to its great SQL support and a large community of PostgreSQL users who are already familiar with the RDBMS it is based on e.g. HERE.
Now, let’s go through the individual scripts and unpack the whole solution piece by piece. The following Python script is using ARM templates to provision Azure resource group and a small Citus database cluster (single coordinator node and two worker nodes).
import adal
from timeit import default_timer as timer
from sys import platform
from msrestazure.azure_active_directory import AdalAuthentication
from msrestazure.azure_cloud import AZURE_PUBLIC_CLOUD
from azure.mgmt.resource.resources.models import (
DeploymentMode,
DeploymentProperties,
Deployment,
)
from azure.mgmt.resource import ResourceManagementClient
from pathlib import Path, PurePosixPath
import json
import os
arm_template_location = PurePosixPath(
"/home/airflow/airflow/dags/azure_citus_db_dag_deps/azure_arm_templates/pg_template.json"
)
arm_template_params_location = PurePosixPath(
"/home/airflow/airflow/dags/azure_citus_db_dag_deps/azure_arm_templates/pg_template_params.json"
)
resource_group = "citus_svr_resource_group"
external_IP = os.popen("curl -s ifconfig.me").readline()
deployment_name = "azure_citus_deployment"
external_ip_params = {
"name": "ClientIPAddress",
"startIPAddress": external_IP,
"endIPAddress": external_IP,
}
admin_login = {"value": "citus"}
admin_passwd = {"value": "your_citus_password"}
template = arm_template_location
params = arm_template_params_location
if template:
with open(template, "r") as json_file_template:
template = json.load(json_file_template)
if params:
with open(params, "r") as json_file_template:
params = json.load(json_file_template)
params = {k: v for k, v in params["parameters"].items()}
params["firewallRules"]["value"]["rules"][1] = external_ip_params
params["administratorLogin"] = admin_login
params["administratorLoginPassword"] = admin_passwd
resource_group_location = params["location"]["value"]
# Tenant ID for your Azure Subscription
TENANT_ID = "123a27e2-7777-4d30-9ca2-ce960d430ef8"
# Your Service Principal App ID
CLIENT = "ae277f4e-882d-4f03-a0b5-b69275046123"
# Your Service Principal Password
KEY = "e7bbf0ed-d461-48c7-aace-c6a4822a123e"
# Your Azure Subscription ID
subscription_id = "1236a74c-dfd8-4b4d-b0b9-a355d2ec793e"
LOGIN_ENDPOINT = AZURE_PUBLIC_CLOUD.endpoints.active_directory
RESOURCE = AZURE_PUBLIC_CLOUD.endpoints.active_directory_resource_id
context = adal.AuthenticationContext(LOGIN_ENDPOINT + "/" + TENANT_ID)
credentials = AdalAuthentication(
context.acquire_token_with_client_credentials, RESOURCE, CLIENT, KEY
)
client = ResourceManagementClient(credentials, subscription_id)
def create_citus_instance(resource_group):
deployment_properties = {
"mode": DeploymentMode.incremental,
"template": template,
"parameters": params,
}
deployment_async_operation = client.deployments.create_or_update(
resource_group, deployment_name, deployment_properties
)
deployment_async_operation.wait()
def main():
print("\nCreating Azure Resource Group...")
start = timer()
client.resource_groups.create_or_update(
resource_group, {"location": resource_group_location}
)
print("Creating Citus Cluster...")
create_citus_instance(resource_group)
end = timer()
time = round(end - start, 1)
print("Total Elapsed Deployment Time = {t} seconds".format(
t=format(time, ".2f")))
if __name__ == "__main__":
main()
This file, called provision_citus_cluster.py, will be called from our Airflow DAG using PythonOperator and is the first step of our pipeline. The instance creation process should take a few minutes to provision and once created we can continue on and generate Citus cluster schema with all the required objects.
In order to populate Citus database with the flat files data we first need to create the desired schema (in this case called tpc_ds) and all the required objects. Even though this process will not be populating all tables (there are only thirteen flat files to be loaded), the script will create all TPC-DS benchmark objects using a simple SQL file containing all necessary DDL statements. The actual SQL file can be viewed and downloaded from my OneDrive folder so to keep this post concise, I will only include the Python script which parses the DDL SQL statements and executes those, creating all required tables. This operation (called from create_citus_schema.py file) will generate a second object in our DAG’s chain of tasks.
from pathlib import Path, PurePosixPath
from sys import platform
from psycopg2.extensions import ISOLATION_LEVEL_AUTOCOMMIT
import psycopg2
import sys
sql_schema = PurePosixPath(
"/home/airflow/airflow/dags/azure_citus_db_dag_deps/sql/tpc_ds_schema.sql"
)
pg_args = {
"user": "citus",
"password": "your_citus_password",
"host": "citussvrgroup-c.postgres.database.azure.com",
"port": "5432",
"database": "citus",
"sslmode": "require",
}
pg_schema_name = "tpc_ds"
def get_sql(sql_schema):
"""
Source operation types from the 'create_sqlite_schema' SQL file.
Each operation is denoted by the use of four dash characters
and a corresponding table DDL statement and store them in a dictionary
(referenced in the main() function).
"""
table_name = []
query_sql = []
with open(sql_schema, "r") as f:
for i in f:
if i.startswith("----"):
i = i.replace("----", "")
table_name.append(i.rstrip("\n"))
temp_query_sql = []
with open(sql_schema, "r") as f:
for i in f:
temp_query_sql.append(i)
l = [i for i, s in enumerate(temp_query_sql) if "----" in s]
l.append((len(temp_query_sql)))
for first, second in zip(l, l[1:]):
query_sql.append("".join(temp_query_sql[first:second]))
sql = dict(zip(table_name, query_sql))
return sql
def build_schema(sql_schema, pg_schema_name, **kwargs):
connection = None
try:
connection = psycopg2.connect(**kwargs)
cursor = connection.cursor()
cursor.execute("SELECT version();")
record = cursor.fetchone()
if record:
connection.set_isolation_level(ISOLATION_LEVEL_AUTOCOMMIT)
cursor.execute(
"CREATE SCHEMA IF NOT EXISTS {s};".format(
s=pg_schema_name)
)
conn_params = connection.get_dsn_parameters()
if conn_params["dbname"] == "citus":
sql = get_sql(sql_schema)
ddls = sql.values()
for s in ddls:
try:
cursor.execute(s)
connection.commit()
except (Exception, psycopg2.DatabaseError) as error:
print("Error while creating nominated database object", error)
sys.exit(1)
else:
raise Exception("Failed to connect to citus database.")
except (Exception, psycopg2.Error) as error:
print("Could not connect to your PostgreSQL instance.", error)
finally:
if connection:
cursor.close()
connection.close()
def main():
build_schema(sql_schema, pg_schema_name, **pg_args)
if __name__ == "__main__":
main()
Up next is our third task which loads text files data into the newly created database. The following Python code (solution file is called load_data_into_citus.py) is responsible for taking each of the staged flat files and copying those into the corresponding tables (in a sequential order).
import psycopg2
import os
import sys
from pathlib import Path, PurePosixPath
from psycopg2.extensions import ISOLATION_LEVEL_AUTOCOMMIT
tpc_ds_files = PurePosixPath(
"/home/airflow/airflow/dags/azure_citus_db_dag_deps/tpc_ds_data/"
)
pg_schema_name = "tpc_ds"
encoding = "iso-8859-1"
pg_args = {
"user": "citus",
"password": "your_citus_password",
"host": "citussvrgroup-c.postgres.database.azure.com",
"port": "5432",
"database": "citus",
"sslmode": "require",
}
def load_data(tpc_ds_files, pg_schema_name, **kwargs):
connection = None
files = [f for f in os.listdir(tpc_ds_files) if f.endswith(".csv")]
try:
connection = psycopg2.connect(**kwargs)
cursor = connection.cursor()
cursor.execute("SELECT version();")
record = cursor.fetchone()
if record:
connection.set_isolation_level(ISOLATION_LEVEL_AUTOCOMMIT)
for file in files:
schema_table_name = pg_schema_name + "." + file[:-4]
with open(
os.path.join(tpc_ds_files, file), "r", encoding=encoding
) as f:
# next(f) # Skip the header row
print(
"Truncating table {table_name}...".format(
table_name=schema_table_name
)
)
cursor.execute(
"TRUNCATE TABLE {table_name};".format(
table_name=schema_table_name
)
)
print(
"Copying file {file_name} into {table_name} table...\n".format(
file_name=file, table_name=schema_table_name
)
)
cursor.copy_from(f, schema_table_name, sep="|", null="")
file_row_rounts = sum(
1
for line in open(
os.path.join(tpc_ds_files, file),
encoding=encoding,
newline="",
)
)
cursor.execute(
"SELECT COUNT(1) FROM {tbl}".format(
tbl=schema_table_name)
)
record = cursor.fetchone()
db_row_counts = record[0]
if file_row_rounts != db_row_counts:
raise Exception(
"Table {tbl} failed to load correctly as record counts do not match: flat file: {ff_ct} vs database: {db_ct}.\
Please troubleshoot!".format(
tbl=schema_table_name,
ff_ct=file_row_rounts,
db_ct=db_row_counts,
)
)
except Exception as e:
print(e)
finally:
if connection:
cursor.close()
connection.close()
def main():
load_data(tpc_ds_files, pg_schema_name, **pg_args)
if __name__ == "__main__":
main()
As you may expect, each one of the aforementioned operations needs to be executed sequentially as there are clear dependencies across each of the steps outlined above i.e. database objects cannot be populated without the schema being created in the first place and likewise, database schema cannot be created without the database instance being present. However, when it comes to SQL queries execution, Citus was designed to crunch large volumes of data concurrently. Our next set of operation will calculate and generate four separate SQL results and stage those as CSV files inside the ‘queries_output’ folder. Please also note that this post does not look into Citus cluster performance and is not concerned with the queries execution times. We are not attempting to tune the way the data is distributed across the nodes or adjust any configuration parameters as the main purpose of this post is to highlight Airflow functionality and design patterns for the above scenarios.
import psycopg2
import os
import sys
import csv
from pathlib import Path, PurePosixPath
sql_queries_file = PurePosixPath("/home/airflow/airflow/dags/azure_citus_db_dag_deps/sql/queries.sql")
sql_output_files_dir = PurePosixPath(
"/home/airflow/airflow/dags/azure_citus_db_dag_deps/queries_output/"
)
pg_schema_name = "tpc_ds"
encoding = "iso-8859-1"
pg_args = {
"user": "citus",
"password": "your_citus_password",
"host": "citussvrgroup-c.postgres.database.azure.com",
"port": "5432",
"database": "citus",
"sslmode": "require",
}
def get_sql(sql_queries_file):
query_number = []
query_sql = []
with open(sql_queries_file, "r") as f:
for i in f:
if i.startswith("----"):
i = i.replace("----", "")
query_number.append(i.rstrip("\n"))
temp_query_sql = []
with open(sql_queries_file, "r") as f:
for i in f:
temp_query_sql.append(i)
l = [i for i, s in enumerate(temp_query_sql) if "----" in s]
l.append((len(temp_query_sql)))
for first, second in zip(l, l[1:]):
query_sql.append("".join(temp_query_sql[first:second]))
sql = dict(zip(query_number, query_sql))
return sql
def main():
query_sql = sql.get(param).rstrip()
query_sql = query_sql[:-1] # remove last semicolon
output_file_name = param.lower() + "_results.csv"
try:
connection = psycopg2.connect(**pg_args)
cursor = connection.cursor()
cursor.execute("SELECT version();")
record = cursor.fetchone()
if record:
sql_for_file_output = "COPY ({0}) TO STDOUT WITH CSV DELIMITER '|';".format(query_sql)
with open(os.path.join(sql_output_files_dir, output_file_name), "w") as output_file:
cursor.copy_expert(sql_for_file_output, output_file)
except Exception as e:
print(e)
finally:
if connection:
cursor.close()
connection.close()
if __name__ == "__main__":
if len(sys.argv[1:]) == 1:
sql = get_sql(sql_queries_file)
param = sys.argv[1]
query_numbers = [q for q in sql]
if param not in query_numbers:
raise ValueError(
"Incorrect argument given. Choose from the following numbers: {q}".format(
q=" or ".join(query_numbers)
)
)
else:
param = sys.argv[1]
main()
else:
raise ValueError(
"Too many arguments given. Looking for <query number> numerical value."
)
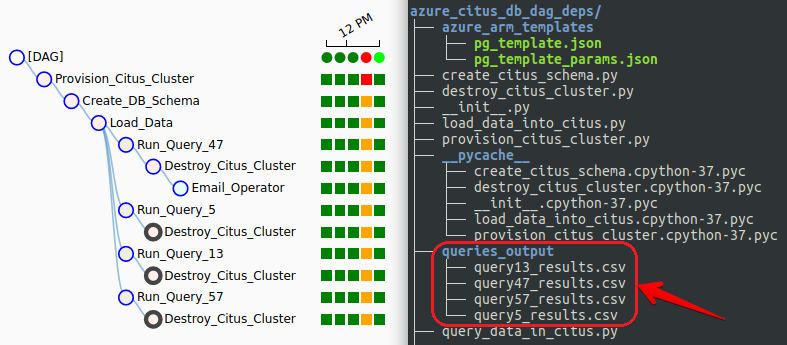
The above code is executed using BashOperator rather than PythonOperator, as each query is run with a parameter passed to the executing script, indicating the SQL query number. Additionally, each query output (the result set) is staged as a CSV file on the local drive so that at the end of the pipeline execution we should end up with four flat files in the queries_output folder.
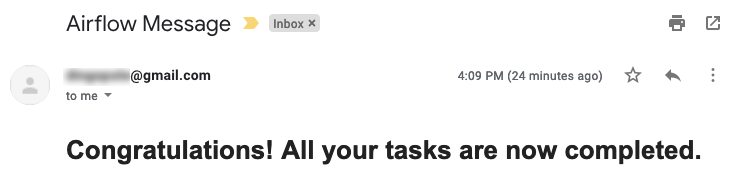
Last but not least, we will initialize the ‘clean-up’ step, where Citus cluster resource group and all its corresponding services will be terminated to avoid incurring additional cost and send an email to a nominated email address, notifying administrator of the successful pipeline execution completion. The following Python code terminates and deletes all resources created in this process.
import adal
from timeit import default_timer as timer
from sys import platform
from msrestazure.azure_active_directory import AdalAuthentication
from msrestazure.azure_cloud import AZURE_PUBLIC_CLOUD
from azure.mgmt.resource.resources.models import (
DeploymentMode,
DeploymentProperties,
Deployment,
)
from azure.mgmt.resource import ResourceManagementClient
resource_group = "citus_svr_resource_group"
# Tenant ID for your Azure Subscription
TENANT_ID = "123a27e2-7777-4d30-9ca2-ce960d430ef8"
# Your Service Principal App ID
CLIENT = "ae277f4e-882d-4f03-a0b5-b69275046123"
# Your Service Principal Password
KEY = "e7bbf0ed-d461-48c7-aace-c6a4822a123e"
# Your Azure Subscription ID
subscription_id = "1236a74c-dfd8-4b4d-b0b9-a355d2ec793e"
LOGIN_ENDPOINT = AZURE_PUBLIC_CLOUD.endpoints.active_directory
RESOURCE = AZURE_PUBLIC_CLOUD.endpoints.active_directory_resource_id
context = adal.AuthenticationContext(LOGIN_ENDPOINT + "/" + TENANT_ID)
credentials = AdalAuthentication(
context.acquire_token_with_client_credentials, RESOURCE, CLIENT, KEY
)
client = ResourceManagementClient(credentials, subscription_id)
def main():
client.resource_groups.delete(resource_group)
if __name__ == "__main__":
main()
There are many ways one can script out email notification dispatch using Python, however, Airflow provides out-of-the-box functionality in the form of EmailOperator. For this demonstration I decided to use my personal Gmail address which required a slight change to the airflow.cfg (Airflow configuration) file as well as creating a Google account App Password. More on the process of creating App Password can be found HERE. The following changes to the Airflow configuration file are required (in addition to DAG-specyfic email task definition) to enable EmailOperator functionality with Gmail-specyfic email address.
smtp_host = smtp.gmail.com smtp_user = your_gmail_email_address@gmail.com smtp_password = your_app_password smtp_port = 587 smtp_mail_from = Airflow
The final piece of the puzzle is the actual DAG file responsible for individual tasks definition as per my previous introductory post to Airflow HERE. The following Python code is responsible for defining DAG arguments, individual tasks and the order they should be executed in. Notice that some Python tasks are executed using BashOperator instead of PythonOperator. That’s because those scripts are run with an argument passed to them using Python sys module argv list, denoting query number which the script is to run. Also, those tasks need to include full path to where the script is located. In contrast, PythonOperator only requires a Python callable to be included.
from airflow import DAG
from airflow.operators.bash_operator import BashOperator
from airflow.operators.python_operator import PythonOperator
from datetime import datetime, timedelta
from airflow.operators.email_operator import EmailOperator
from azure_citus_db_dag_deps import create_citus_schema
from azure_citus_db_dag_deps import load_data_into_citus
from azure_citus_db_dag_deps import provision_citus_cluster
from azure_citus_db_dag_deps import destroy_citus_cluster
default_args = {
'owner': 'Airflow',
'depends_on_past': False,
'start_date': datetime(2019, 6, 1),
'email': ['airflow@example.com'],
'email_on_failure': False,
'email_on_retry': False,
'retries': 0,
# 'retry_delay': timedelta(minutes=5),
# 'queue': 'bash_queue',
# 'pool': 'backfill',
# 'priority_weight': 10,
# 'end_date': datetime(2016, 1, 1),
}
with DAG('Azure_Citus_Database_Job', default_args=default_args, schedule_interval=None) as dag:
provisioning_citus_cluster = PythonOperator(
task_id="Provision_Citus_Cluster",
python_callable=provision_citus_cluster.main)
creating_citus_schema = PythonOperator(
task_id="Create_DB_Schema", python_callable=create_citus_schema.main)
loading_csv_data = PythonOperator(
task_id="Load_Data", python_callable=load_data_into_citus.main)
executing_sql_query_5 = BashOperator(
task_id="Run_Query_5", bash_command='python ~/airflow/dags/azure_citus_db_dag_deps/query_data_in_citus.py Query5'
)
executing_sql_query_13 = BashOperator(
task_id="Run_Query_13", bash_command='python ~/airflow/dags/azure_citus_db_dag_deps/query_data_in_citus.py Query13'
)
executing_sql_query_47 = BashOperator(
task_id="Run_Query_47", bash_command='python ~/airflow/dags/azure_citus_db_dag_deps/query_data_in_citus.py Query47'
)
executing_sql_query_57 = BashOperator(
task_id="Run_Query_57", bash_command='python ~/airflow/dags/azure_citus_db_dag_deps/query_data_in_citus.py Query57'
)
destroying_citus_cluster = PythonOperator(
task_id="Destroy_Citus_Cluster", python_callable=destroy_citus_cluster.main)
emailing_operator = EmailOperator(
task_id="Email_Operator",
to="your_gmail_email_address@gmail.com",
subject="Airflow Message",
html_content="""<h1>Congratulations! All your tasks are now completed.</h1>"""
)
provisioning_citus_cluster >> creating_citus_schema >> loading_csv_data >> executing_sql_query_5 >> destroying_citus_cluster >> emailing_operator
provisioning_citus_cluster >> creating_citus_schema >> loading_csv_data >> executing_sql_query_13 >> destroying_citus_cluster >> emailing_operator
provisioning_citus_cluster >> creating_citus_schema >> loading_csv_data >> executing_sql_query_47 >> destroying_citus_cluster >> emailing_operator
provisioning_citus_cluster >> creating_citus_schema >> loading_csv_data >> executing_sql_query_57 >> destroying_citus_cluster >> emailing_operator
Now that we have everything in place we can kick this DAG off and wait for the pipeline to go through its individual execution steps, concluding with the queries output files staged in the nominated folder as well as the email sent to the Gmail address we specified.
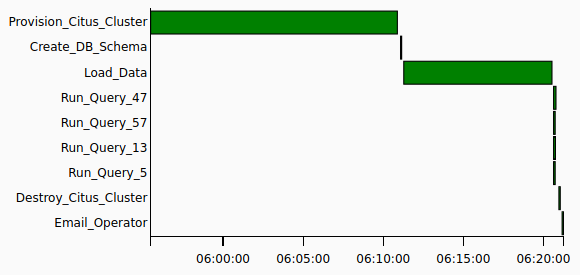
When finished, Airflow can provide us with the Gantt chart breakdown on how long individual tasks took which helps with pinpointing slow-running processes and gives us a good overview of time consumed across each step.
Additionally, we can now see the DAG tree view completion status (green = success, red = failure) for each run and each step, the four output files staged in the nominated directory with expected queries result sets persisted and finally, the email notification sent out on pipeline successful completion.
Conclusion
This concludes this short series outlining some of the core functionality of Airflow workflow management system. From the short time I spent with Airflow it seems like it fills the void for a robust, open-source, configuration-as-code platform which can be used across many different application, not just ETL/ELT e.g. automating DevOps operations, machine learning pipelines, scheduler replacement etc. Its documentation is passable, it has a fairly mild learning curve, it’s fairly extensible and scalable, has a good management interface and its adoption rate, particularly in the startup community, is high as it is slowly becoming the de facto platform for programmatic workloads authoring/scheduling. Data engineering field has been changing rapidly over the last decade, competition is rife and I’m not sure whether Airflow will be able to sustain its momentum in years to come. However, given its current rise to stardom, easily superseding tools such as Apache Oozie or Luigi, anyone in the field of Data Engineering should be at least familiar with it.



Never thought Polybase could be used in this capacity and don't think Microsoft advertised this feature (off-loading DB data as…